This document describes the work done on the AICE Working Group demonstrator with the AURA use case. This project constitutes the first iteration of the AICE OpenLab and intends to demonstrate the benefits of a shared, common platform to collaboratively work on AI workflows.
The Eclipse AI, Cloud & Edge (AICE) Working Group is a special interest working group hosted at the Eclipse Foundation, where participants discuss and work on AI, cloud & edge ecosystems to innovate and grow with open source. The aim of the AICE Working Group is to accelerate the adoption of AI, cloud & edge technologies and standards through the provision and operation of a collaborative work and test environment for its participants, engagement with research and innovation initiatives, and the promotion of open source projects to AI, cloud & edge developers.
The AICE OpenLab has been initiated to provide a common shared platform to test, evaluate and demonstrate AI workflows developed by partners. This enables an open collaboration and discussion on AI solutions, and fosters portability and standardisation. The AICE OpenLab is currently working on two use cases: AURA, as described in this document, and Eclipse Graphene, a general-purpose scheduler for AI workflows.
More information:
AURA is a non-profit French organisation that designs and develops a patch to detect epileptic seizures before they happen and warns patients ahead for safety purposes. For this AURA is creating a multidisciplinary community integrating open source and open hardware philosophies with the health and research worlds. The various partners of the initiative (patients, neurologists, data scientists, designers) each bring their experience and expertise to build an open, science-backed workflow that can actually help the end-users. In the end, this device could be a life-changer for the 10 million people with drug-resistant epilepsy worldwide.
More information:
The epileptic seizure is the cornerstone for the management of the disease by health professionals. A precise mapping of seizures in daily life is a way to better qualify the effectiveness of treatments and care. This is a first step towards the forecasting of epileptic seizures which would allow people to better have control over their epilepsy and regain autonomy in their daily lives.
There are a myriad of different forms and origins of epilepsy and epileptic seizures. The symptoms and physical signs broadly differ according to each patient: research has been conducted on electroencephalograms (EEGs), electrocardiograms (ECGs), movement detection, electrodermal activity, and even using dogs. As a result it is impossible — as of today at least — to draw a generic-purpose diagnostic or prediction method.
Machine Learning (ML) algorithms have been extensively used in the recent years to tackle this variability across patients and draw viable seizure detection and forecasting. Generally speaking, ML methods rely on a (large) set of examples, in our case datasets of ECGs with their associated seizure annotations, to predict specific outputs (epileptic seizures) on new input data (e.g. live ECGs). Neurophysiologists use a visualisation tool like Grafana to enter the annotations and define time ranges as either normal activity (noise) or epileptic activity (seizures), and store them in dedicated (.tse_bi) files.
These annotations are used as a reference dataset for training various ML models. Available datasets are usually split so as to set one part for the training and another one to verify the trained model. A typical workflow is to then try to predict epileptic seizures according to an ECG signal and check if the human annotations confirm the seizure.
More information:
We started from the workflow already developed by the AURA data scientists. As usual the first step is to prepare the data before using it (cleaning, selection and extraction of features, and formatting). The resulting dataset is subsequently fed to a ML model to predict future seizures.
The data inputs of the workflow come from two different file types:
.tse_bi files with a 1-to-1 association with the EDF signal files.The data preparation step is achieved through a series of Python scripts developed by the AURA scientists, which extract the rr-intervals (i.e. the time between two heart beats), cardiac features and annotations, then build a simplified dataset that can be used to train a Random-Forest algorithm:
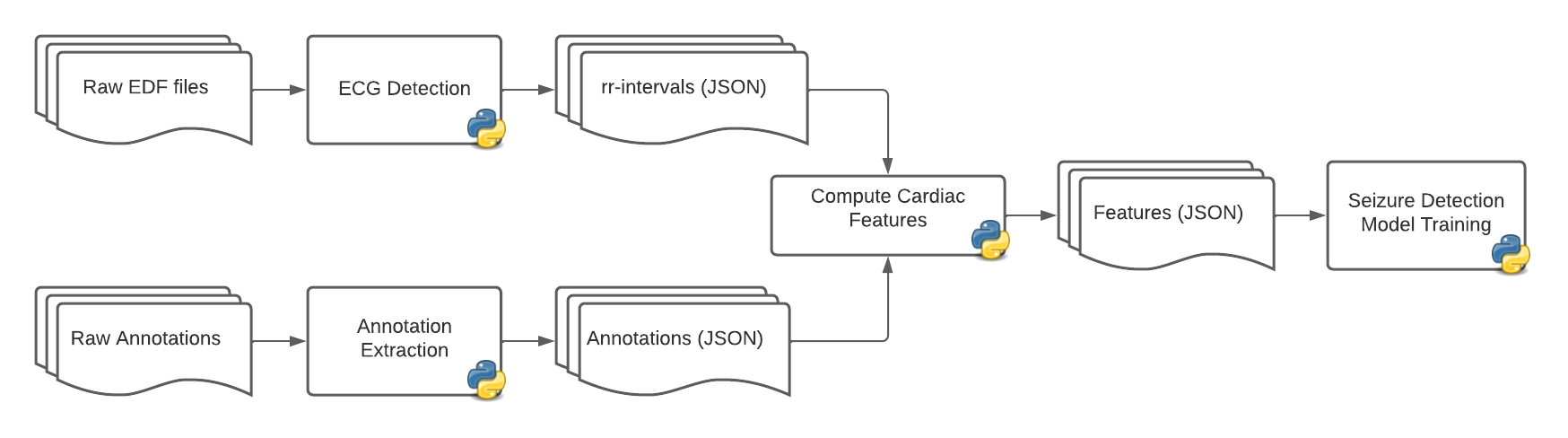
There are many parameters involved in the process, with some of them having a huge impact on performance, like the time window for the rr-interval. In order to fine-tune these parameters one needs to try and run various combinations, which is not practical and can be prohibitive with long executions.
More information:
In this context, our first practical goal was to train the model on a large dataset of ECGs from the Temple University Hospital (TUH). The TUH dataset is composed of EDF files recording the electrocardiogram signal, along with their annotation files that classify the time ranges as either noise or as an epileptic seizure. The full dataset has 5600+ EDF files and as many annotations, representing 692 patients, 1074 hours of recordings and 3500+ seizures. Its size on disk is 67GB.
AI-related research and development activities, even if they rely on smaller datasets for the early stages of the set up, require a more complete dataset to run when it comes to the fine-tuning and exploitation of the model. The TUH database was not used often with the previous AURA workflow, as its full execution would take more than 20 hours on the developer’s computers. Executions often failed because of wrong input data, and switching to more powerful computers was difficult because of the complex setup.
Established objectives of the project were to:
More information:
Considering the above situation and objectives, we identified four area of improvements:
One key aspect of the work achieved was to make the AI workflow easy to run anywhere, from the researcher’s computers to our Kubernetes cluster. This implies to have a set of scripts and resources to automatically build a set of Docker images for each identified step of the process. On top of drastically improving portability, it also means that the very same workflow can be identically reproduced on different datasets.
We developed three Docker images to easily execute the full workflow or specific steps:
docker run -v $(pwd)/data/:/data bbaldassari/aura_dataprep bash /aura/scripts/run_bash_pipeline.shdocker-compose -f docker-compose-dataprep.yml up -dWe decomposed the workflow into a sequence of steps that could be encapsulated and executed sequentially, i.e. where each step needs the output of the previous step to start. On steps that allow it (e.g. data processing) we can also run several containers in parallel. The resulting architecture is shown below:
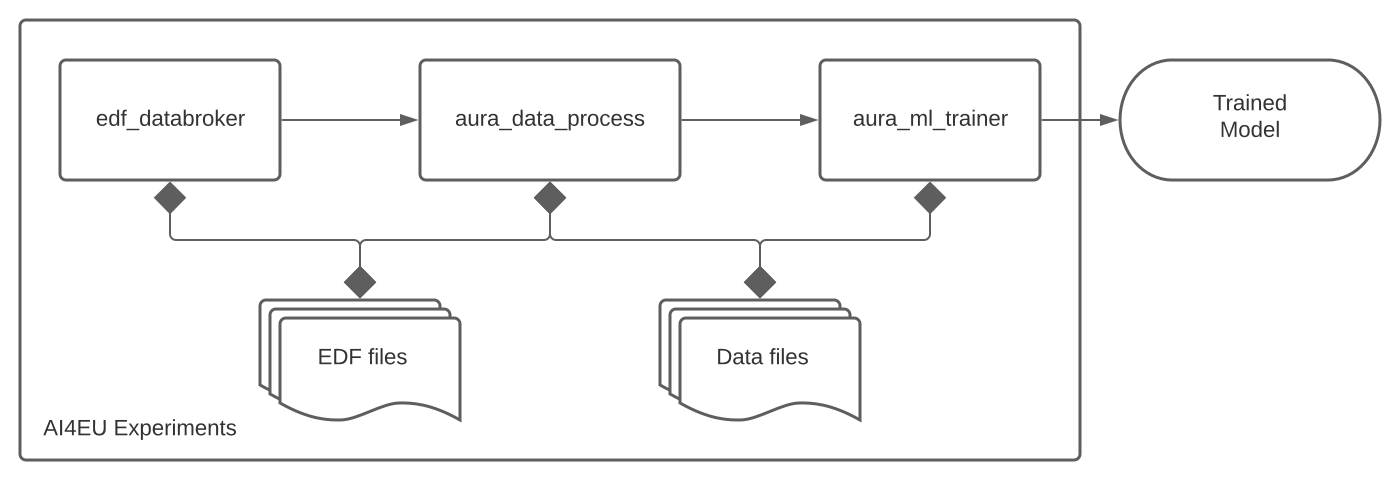
In this diagram, the following containers are defined:
All containers work on the same set of directories:
The images have been imported into our instance of AI4EU Experiments for further dissemination and collaboration.
Another step was to refactor the scripts to identify and remove performance bottlenecks. Things that work well on a small dataset can become unusable on a larger scale. By running it on larger datasets, up to thousands of files (i.e. the TUH dataset) we encountered unexpected cases and fixed them along the way. We now have a set of scripts that 1. can run on the entire TUH dataset (67GB) without major issue, and 2. is compatible with the two data formats most used by the AURA researchers: TUH and La Teppe.
The performance gain enabled us to run more precise and resource-consuming operations in order to refine the training. For example we modified the length of the sliding window when computing the rr-intervals from 9 seconds to 1 second, which generates a substantial amount of computations while seriously improving predictions from the ML training.
We identified atomic steps that could be executed independently and built them as parallel execution jobs. As an example, the cleaning and preparation of data files can be executed simultaneously on different directories to accelerate the overall step. By partitioning the dataset in subsets of roughly 10GB and running concurrently 6 data preparation containers we went down from almost 17 hours to 4 hours on the same reference host.
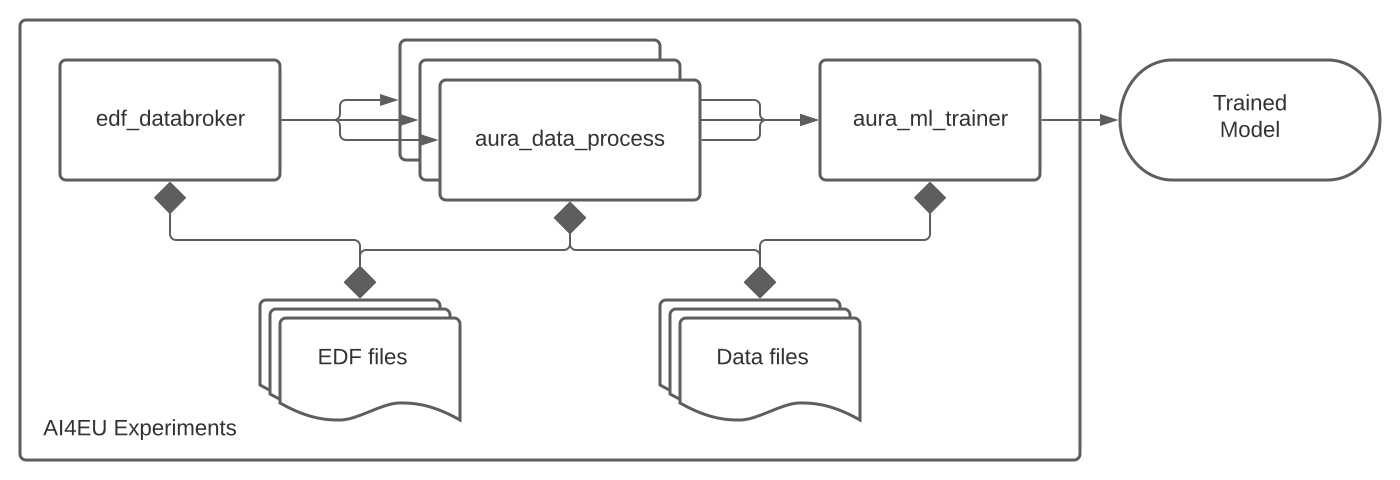
Also by being able to run the process everywhere, we could execute it on several hardwares with different capabilities. This allowed us to check (and fix) portability while getting a better understanding of the resource requirements of each step. We targeted three different hosts for our performance benchmark:
The following plot shows the evolution of performance in various situations:
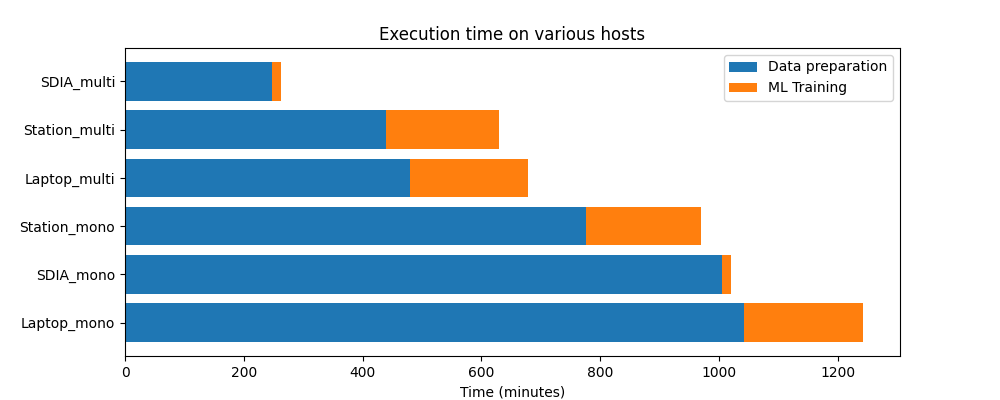
We could identify different behaviours regarding performance. The data preparation step relies heavily on IOs, and improving the disk throughput (e.g. SSD + NVMe instead of a classic HDD) shows a 30% gain. The ML training on the other hand is very CPU- and memory- intensive, and running it on a node with a large number of threads (e.g. 48 in our case) brings a stunning 10x performance improvement compared to a laptop equipped with an Intel i7.
AURA uses Grafana to display the ECG signals and the associated annotations, both for the creation of annotated data sets and for their exploitation. In order to build this workflow we need to import the rr-intervals files and their associated annotations in a PostgreSQL database, and configure Grafana to read and display the corresponding time series.
An example of rr-interval plot with the associated annotations (blue/red bottom line) is shown below:
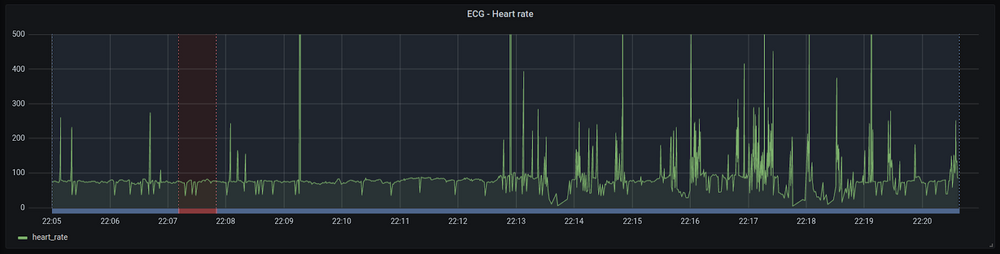
The process of importing the rr-intervals and annotations is time- and resource- consuming, so we decided to apply the same guidelines as for the training workflow and built a dedicated container for the mass import of ECG signals with their annotations. By partitioning the dataset and setting up multiple containers we are able to run several import threads in parallel, thus massively improving the overall performance of the import. It enabled us to:
It is also very important to interpret visually and discuss the outcomes of the AI-based seizure detector with the healthcare professionals in order to build trust and assess limitation of the algorithm. Having an easy way to import ECGs to easily visualise and annotate them is a major benefit in this context, especially in healthcare centers where teams do not always have the resources and knowledge to set up a complex software stack. We are now working on a database dump that will enable end-users to import specific datasets into their own Postgres / Grafana instance in a few clicks, thus fostering the usage and research on open datasets.
The work done by the AURA researchers and data scientists on ECGs had been organised in a bunch of GitHub repositories, with different people using different tools and structures. The first step was to identify the parts required to run the complete workflow, and extract them from the various repositories and branches to build a unified structure. The requirements of this repository structure are:
Building upon the current resources in use at AURA for Ai workflow, the following directory structure was adopted:
├── data => Data samples for tests
├── graphene => All Docker images
│ ├── aura_dataprep => - Data processing Docker image
│ ├── aura_ml_trainer => - ML training Docker image
│ └── ...
├── resources => Documentation, images..
├── scripts => Repo-related scripts for builds, integration..
├── src => AI-related scripts and source code
│ ├── domain
│ ├── infrastructure
│ └── usecase
└── tests => Tests for scripts and source code
We defined and enforced a contributing guide, making tests and documentation mandatory in the repository. We also set up a Travis job to execute the full python test suite at every commit, and made it visible through a badge in the repository’s README. Regarding Git we used a simple Git workflow to maintain a clean branching structure. The newly agreed development process definitely helped clean up the repository. Each time a set of scripts was added, we knew exactly where they should go and how to reuse them in the overall workflow.
Once the new Docker images are built and pushed to a Docker registry, they can be pulled from any computer in order to run the full workflow without any local install or specific knowledge. The provided docker-compose files will automatically pull the required images and execute the full workflow on any dataset and on any host. An example is proposed to fine-tune and run the multi-containers setup easily. For the teams at AURA, it means they can now run their workflows on any type of hosting provided by their partners.
We also installed a fresh instance of AI4EU Experiments on our dedicated hardware for the onboarding of the models, and plan to make stable, verified images available on the marketplace in the upcoming months.
The major performance gain was achieved by setting up dedicated containers to run atomic tasks (e.g. data preparation, visualisation imports) in parallel. Most computers, both in the lab and for high-end execution platforms, have multiple threads and enough memory to manage several containers simultaneously, and we need to take advantage of the full computing power we have. Another major gain was obviously to run the process on a more powerful system, with enough memory, CPUs and disk throughput.
All considered we were able to scale down the full execution time on the TUH dataset from 20 hours on the lab’s laptop to roughly 4 hours in our cluster.
The new repository has a sound and clean structure, with passing tests, a complete documentation to exploit and run the various steps, and has everything needed for further developments. All scripts are stored under the src/ directory and are copied to the docker images during the build, thus always relying on a single source of tested truth.
Furthermore, the automatic building of containers for multiple execution targets (Airflow, Docker, Kubernetes) can easily be reproduced. As a result the new, improved structure will be reused and is set to become the reference implementation for the next developments.
It has been a fantastic collaborative work, building upon the expertise of the AURA data scientists and AICE MLOps practitioners to deliver exciting and pragmatic outcomes. The result is a set of optimised, reliable processes, with new perspectives and possibilities, and a better confidence in the developed pipeline. All actors learned a lot and the sequels of the work will be replicated in the forthcoming projects in both teams.
Besides the team benefits, the project itself hugely benefited from the various improvements and optimisation. It is now very easy to run the full stack on different datasets for development, and the new container deployment method will be extended to partners and healthcare centers (L’Institut La Teppe).
We identified a few areas of improvement, though. One aspect that we lacked in this experience was a precise benchmarking process and framework for the various steps, at each optimisation round. We are currently working on a monitoring solution based on Prometheus, Node exporter and Grafana to solve the issue, and we will be publishing soon a more detailed report on the performance gains.